Intracellular IRF5 Dimerization Assay
Cherrie D. Sherman and Betsy J. Barnes
Center for Autoimmune Musculoskeletal and Hematopoietic Diseases, Feinstein Institutes for Medical Research, Manhasset, New York, USA
For correspondence: cherriesherman.phd@gmail.com; bbarnes1@northwell.edu
Abstract
The intracellular interferon regulatory factor 5 (IRF5) dimerization assay is a technique designed for measuring molecular interaction(s) with endogenous IRF5. Here, we present two methods that detect endogenous IRF5 homodimerization and interaction of endogenous IR5 with cell penetrating peptide (CPP) inhibitors. For detection of endogenous IRF5 dimers, THP-1 cells were incubated in the presence or absence of IRF5-targeted CPP (IRF5-CPP) inhibitor for 30 minutes and subsequently stimulated with R848 for 1 hour. Cell lysates were separated by native-polyacrylamide gel electrophoresis (PAGE) and IRF5 dimers detected by immunoblotting with IRF5 antibodies. For detection of endogenous IRF5 with FITC-labeled IRF5-CPP, the in-cell fluorescence resonance energy transfer (FRET) assay was used. THP-1 cells were left untreated or treated with FITC-IRF5-CPP conjugated inhibitors for 1 hour. Cells were fixed, permeabilized, stained with anti-IRF5 and TRITC-conjugated secondary antibodies. The transfer of fluorescence was measured and calculated as FRET units. These methods provide rapid and accurate assays to detect IRF5 molecular interactions.
Keywords: IRF5, Native-PAGE, polyacrylamide gel electrophoresis, FRET, dimerization, molecular interaction, IRF5-CPP
Background
Interferon regulatory factor 5 (IRF5) is a transcription factor that regulates pathogen-induced innate and acquired immune responses downstream of Toll-like receptor (TLR), retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), and B cell receptor (BCR). IRF5 has been implicated in the pathogenesis of systemic lupus erythematosus due to its role in regulating the expression of proinflammatory cytokines such as IFN-α, IL-6, TNF-α, and IL-12 and pathogenic autoantibody production. In an unstimulated condition, IRF5 is generally localized in the cytoplasm as a monomer. Activation of the above receptors leads to signaling cascades. IRF5 undergoes post-translational modification, which eventually leads to homodimerization, a critical event prior to nuclear translocation. Here, we describe two methods designed for detecting IRF5 molecular interactions.
In our study, the native-PAGE method was used to detect endogenous IRF5 homodimers. THP-1 cells were incubated with or without (1 and 10 µM) IRF5-CPP inhibitors for 30 minutes, and subsequently stimulated with 1 µM of R848 (a TLR7 ligand) or left unstimulated for 1 hour. Cell lysates were run on native polyacrylamide gel electrophoresis (PAGE) and IRF5 dimerization was detected by immunoblotting with IRF5 and HRP-conjugated secondary antibodies. The intracellular fluorescence resonance energy transfer (FRET) assay was used to detect binding of endogenous IRF5 to FITC-IRF5-CPP conjugated inhibitors through a FRET signal. FRET is a technique designed for detecting molecular interaction in which the excited molecule (the donor) transfers non-radiative energy to another molecule (the acceptor) within a distance of ~1–10nm. THP-1 cells were incubated with or without (1 and 10 µM) IRF5-CPP inhibitors for 1 hour. The untreated and FITC-IRF5-CPP- treated THP-1 cells were fixed, permeabilized, stained with anti-IRF5, anti-IRF3, or anti-IRF7 (other IRF family members) antibodies and TRITC secondary antibodies. Cell-associated fluorescence was measured on a BioTek Synergy Neo2 at 525 nm upon excitation at 488 nm (E1), at 600 nm upon excitation at 540 nm (E2), and at 600 nm upon excitation at 488 nm (E3). The transfer of fluorescence was calculated as FRET unitsas follows: FRET unit = (E3 both − E3 none ) − ([E3 TRITC − E3 none ) × (E2 both /E2 TRITC ]) − ([E3 FITC − E3 none ] × [E1 both /E1 FITC ]). The different fluorescence values (E) were measured on unlabeled cells (E none ) or cells labeled with FITC (E FITC ) and TRITC (E TRITC ).
These techniques can be broadly applied to evaluate intracellular molecular interactions in living cells through pharmacological and molecular studies. They provide a rapid and reliable method of screening molecular interactions, which require standard equipment that are readily available in almost every laboratory.
I. Native-PAGE (Polyacrylamide Gel Electrophoresis)
Graphic abstract
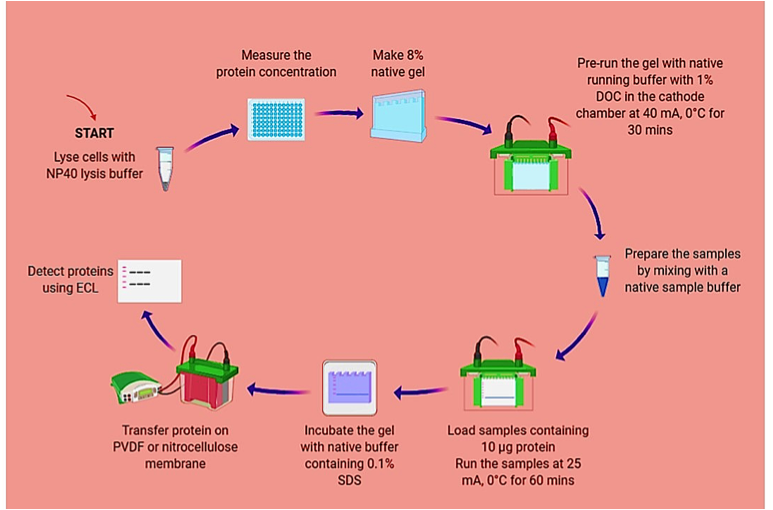 Figure 1. Overview of Native-PAGE method designed for detecting endogenous IRF5 homodimer
Figure 1. Overview of Native-PAGE method designed for detecting endogenous IRF5 homodimer
Materials and Reagents
1. ThP1 (ATCC TIB-202)
2. Cell penetrating peptides (inhibitors) (Hoffman-LaRoche, Patent # WO2014001229A2)
3. Resiquimod (R848) (SigmaAldrich, Cat # SML0196)
4. Sodium deoxycholate (DOC) (Thermofisher Cat # 89904)
5. DC Protein Assay (BioRad, Cat # 5000111)
6. BCA Protein Assay (ThermoFisher, Cat # 23227)
7. Anti-IRF3 (Abcam, Cat # ab76409)
8. Anti-IRF5 (Cell Signaling Tech, Cat # cs3257 or cs13496)
9. Anti-IRF7 (Cell Signaling Tech, Cat # cs4920)
10. Horseradish peroxidase (HRP)–conjugated monoclonal anti–β-actin (Cell Signaling Tech, Cat
# cs5125)
11. HRP-conjugated secondary antibody (Cell Signaling Tech, Cat # cs7074)
12. NP40 Cell Lysis Buffer (Invitrogen, Cat # FNN0021)
13. Phenylmethylsulfonyl fluoride (PMSF) (SigmaAldrich, Cat # 10837091001)
14. Protease Inhibitor (SigmaAldrich, Cat. # P-2714)
15. Native Gel Running Buffer, Tris-Glycine Buffer 10× Concentrate, pH 8.3 (SigmaAldrich, Cat #
T4904-1L)
16. Glycerol (SigmaAldrich, Cat # G9012)
17. Deoxycholate (DOC) (Thermofisher, Cat # 89904)
18. Bromophenol blue (SigmaAldrich, Cat # B5525)
19. 30% Acrylamide/Bis Solution, 37.5:1, 500 ml (BioRad, Cat # 161-0158)
20. Resolving Gel Buffer, 1.5 M Tris-HCl, pH 8.8, 1L (BioRad, Cat # 161-0798)
21. Stacking Gel Buffer, 0.5 M Tris-HCl, pH 6.8, 1L (BioRad, Cat # 161-0799)
22. SDS Solution, 10% (w/v), 250 ml (BioRad, Cat # 161-0416)
23. WesternBright ECL HRP Substrate (Advansta, Cat # K-12045-C20)
Equipment
1. ChemiDoc MP Imaging System (Bio-Rad, 17001402)
2. Mini-Transblot Cell and PowerPac Basic Power Supply (Bio-Rad, 1703989)
3. Mini-Protean Tetra Vertical Electrophoresis Cell for Mini Precast Gels (Bio-Rad, 1658005)
4. Biotek Synergy Neo2 (Biotek, Cat # BTNEO2) or any similar plate reader
Procedure
A. R848-induced IRF5 homodimerization and inhibition of IRF5 homodimerization by CPPs in ThP1
1. Pre-treat ThP1 cells (6-10x106) with IRF5-CPPs (1 and 10 µM) for 30 mins and subsequently treat cells with R848 for 1 hour.
2. Harvest treated cells and transfer in to a 15 ml tube.
3. Centrifuge at 400xg, 4°C for 5 min. Aspirate supernatant without disturbing the cell pellet.
4. Wash cells with PBS and centrifuge at 400xg, 4°C for 5 min. Aspirate PBS without disturbing the cell pellet.
B. Cell Lysis and Protein Concentration Measurement
1. Prepare cell lysis buffer by adding 1 mM PMSF and 500 mL of 10x protease inhibitor cocktail immediately prior to use. (NP40 lysis buffer, manufacturers instruction: http://tools.thermofisher.com/content/sfs/manuals/FNN0021_Rev%200908.pdf)
2. Lyse the cell pellet in cell lysis buffer for 30 minutes, on ice, and vortex every 10-minutes. The volume of cell lysis buffer depends on the cell number and expression of target protein.
3. Transfer the extract to microcentrifuge tubes and centrifuge at 8000xg for 10 min at 4°C.
4. Aliquot the clear lysate to clean microfuge tubes. These samples are ready for BCA or DC protein assay. Lysates can be stored at -80 °C. Avoid multiple freeze/thaws.
5. Measure the protein concentration using BCA or DC protein assay as per kit instruction. Keep samples on ice to prevent denaturation.
C. 8% Native Polyacrylamide Gel Hand Casting
Make 8% Native Gel (as per BioRad Casting Instruction) http://www.bio-rad.com/webroot/web/pdf/lsr/literature/4110106B.pdf
Note: Substitute SDS with Distilled Deionized Water. SDS is a known amphipathic surfactant that denatures proteins. Do NOT use SDS except for gel incubation post-native PAGE.
D. Native-PAGE and Immunoblotting
1. Put pre-running native running buffer (25 mM Tris and 192 mM glycine, pH 8.3) in the freezer until they achieved 0 °C temperature. Make sure that there are no ice crystals prior to pouring in to the electrophoresis chamber unit.
2. Pre-run native gel with native running buffer with 1% DOC in the cathode chamber, and native running buffer in the anode chamber at 40 mA, 0°C for 30 min. Keep electrophoresis unit on ice and maintain 0°C temperature.
3. Prepare samples for native gel electrophoresis by mixing the sample with native sample buffer. Final protein concentration per sample is 10 µg. Do NOT heat the samples (heating will denature the proteins which will lead to dissociation of the dimer).
4. Add fresh cold native running buffers in the anode and cathode chambers (with 1% DOC in the cathode).
5. Load the ladder and samples (protein: 10 µg) per well.
6. Electrophorese samples at 25 mA, 0 °C, for 60 min.
7. Incubate gel in native running buffer containing 0.1% SDS for 30 min.
8. Proceed to protein transfer on PVDF or nitrocellulose membrane.
9. After transfer, proceed to Western Blot.
10. Detect proteins using ECL.
The resulting immunoblot (Figure 2) shows the endogenous IRF5 dimer, monomer and beta-actin (loading control).
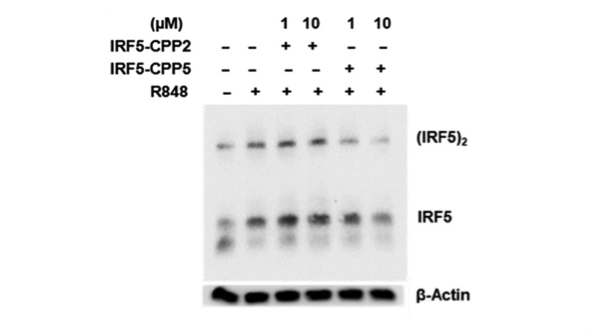
Figure 2. Native-PAGE. On lane 1 control – untreated ThP1, lane 2 treated with R848, lane 3 treated with 1 µM CPP2 and R848, treated with 10 µM CPP2 and R848, lane 5 treated with 1 µM CPP5 and R848, and lane 6 treated with 10 µM CPP5 and R848.
Recipe
A. Native Sample Buffer
1. To make 10 ml of Native Sample Buffer, mix 15% glycerol (v/v), 1% DOC (w/v), and 1.25 ml
of 0.5 M Tris-Cl, pH 6.8 and 0.1% bromophenol blue (w/v) and deionized water.
II. In-cell Fluorescence Resonance Energy Transfer (FRET)
Materials
1. ThP1 (ATCC TIB-202)
2. Cell penetrating peptides (inhibitors) (Hoffman-LaRoche, Patent # WO2014001229A2)
3. Resiquimod (R848) (SigmaAldrich, Cat # SML0196)
4. Foxp3 Fixation/Permeabilization Buffer (Invitrogen, Cat # 00-5523-00)
5. Permeabilization Buffer (Invitrogen, Cat # 00-5523-00)
6. IRF5 antibody (Abcam, Cat # ab124792)
7. TRITC antibody (Abcam, Cat # ab6718)
Equipment
1. Biotek Synergy Neo2 (Biotek, Cat # BTNEO2) or any similar plate reader
Procedure
A. Buffer and Solution Preparation
1. Prepare fresh Foxp3 Fixation/Permeabilization working solution by mixing 1 part of Foxp3 Fixation/Permeabilization concentrate with 3 parts of PBS containing 2% BSA. For 3-5 million cells, 1 ml of the working solution is required for each sample.
2. Prepare 1x working solution of Permeabilization Buffer by mixing 1 part of 10x Permeabilization Buffer with 9 parts of distilled water. 1 ml of the working solution is required for each sample.
B. Detection of IRF5 homodimer-CPP interaction in ThP1
1. For intracellular FRET of FITC-CPP and endogenous IRF5, incubate ThP1 cells with IRF5-CPPs (1 and 10 µM) for 30 min and subsequently treat cells with R848 for 1 h.
2. Harvest the treated cells and transfer in to a 5 ml tube.
3. Centrifuge at 400xg, 4°C for 5 mins. Aspirate supernatant without disturbing the cell pellet.
4. Wash cells with 3 ml PBS and centrifuge at 400xg, 4°C for 5 min. Aspirate PBS without disturbing the cell pellet.
5. Add 1 ml of Foxp3 Fixation/Permeabilization working solution to each tube and pulse vortex.
6. Incubate for 30-60 min at room temperature or for up 18 h at 2-8 °C.
7. Cells should have settled in the bottom after 18 h. Carefully aspirate supernatant. For short incubation, spin at 400xg for 5 min.
8. Add 1 ml of 1X Permeabilization Buffer to each tube and incubate for 30 min at room temperature.
9. Spin at 400xg for 5 min.
10. Resuspend pellet in 50 µl residual volume of 1x Permeabilization Buffer.
11. [Optional] Block with 10% BSA by adding 5 µl directly to each tube. Incubate for 15 mins at room temperature.
12. Without washing, add the recommended amount of directly pre-conjugated antibody for detection of intracellular antigen and incubate for at least 30 mins at room temperature. Protect from the light.
13. Wash with 3 ml of PBS. (If secondary antibody is required, repeat steps 9-12)
14. Spin at 400xg for 5 min. Aspirate supernatant with only 30 µl residual volume.
15. Add 30 µl of 4% PFA and resuspend stained cells. Protect from the light.
16. Transfer samples on a 98-well plate. Prior to reading, add PBS to a final volume of 150-200 µl. NOTE: Make sure you have the same number of cells per well.
17. Measure intracellular fluorescence on Biotek Synergy Neo2 (Bioteck, VT, USA) or any similar plate reader at 525 nm upon excitation at 488 nm (E1), at 600 nm after excitation at 540 nm (E2), and 600 nm after exctitation at 488 nm (E3).
Set up the machine with 3 spectra:
FITC 488 excitation 525 emission
TRITC 540 excitation 600 emission
FRET 488 excitation 600 emission
18. The transfer of fluorescence was calculated as FRET units as follows: FRET unit = (E3c− E3none) − ([E3TRITC− E3none) × (E2none/E2TRITC]) − ([E3 FITC− E3none] × [E1both/E1TRITC ]). The different fluorescence values (E) were measured on unlabeled cells (Enone) or cells labeled with FITC (EFITC) and TRITC (ETRITC).
Acknowledgments
Funding: This work was supported and funded by F. Hoffmann–La Roche, the Lupus Research Alliance (to B.J.B.), Department of Defense CDMRP Lupus Research Program W81XWH-18-1-0674 (to B.J.B.), NIH AR065959 (to B.J.B.), and EMD Serono Research and Development Institute Inc.
Competing interests
Disclosures: J.A.D., S.-L.T., and D.S. are inventors on patent application US20160009772A1 assigned to F. Hoffmann–La Roche AG. Application status abandoned as of 12 May 2019 as a matter of public record. Financial disclosures related to companies: G.C., C.-C.S., J.Q., M.D., and J.A.D. are employees of EMD Serono Research and Development Institute Inc. S.H. is employee of BMS. F.M. is the author of patent “Cell penetrating peptides & methods of identifying cell penetrating peptides” (WO2014001229A2) filed by F. Hoffmann–La Roche. J.A.D., N.F., A.F.H., K.-S.H., F.M., D.S., and S.-L.T. are authors of patent “Cell penetrating peptides which bind IRF5” (US20160009772A1) filed by Hoffmann–La Roche Inc.
References
1. Banga, J., Srinivasan, D., Sun, C-C., Thompson, C.D., Milletti, F., Huang, K.-S., Hamilton, S., Song, S., Hoffman, A. F., Qin, Y. G., Matta, B., LaPan, M., Guo, Q., Lu, G., Li, D., Qian, H., Bolin, D. R., Liang, L., Wartchow, C., Qiu, J., Downing, M., Narula, S., Fotouhi, N., DeMartino, J. A., Tan, S.-L., Chen, G., Barnes, B. J. (2020). Inhibition of IRF5 cellular activity with cell-penetrating peptides that target homodimerization. Sci Adv 6(eaay1057): 1-16.
2. Ujlaky-Nagy, L., Nagy, P., Szollosi, J., Vereb, G. (2018). Flow cytometric FRET analysis of protein interactions. Methods Mol Biol. 1678: 393-419.