往期刊物2025
卷册: 15, 期号: 7
生物化学
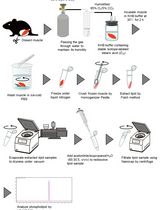
Ex Vivo Measurement of Stable Isotope-Labeled Fatty Acid Incorporation Into Phospholipids in Isolated Mice Muscle
离体条件下稳定同位素标记脂肪酸在分离小鼠骨骼肌磷脂中的掺入测定
生物信息学与计算生物学
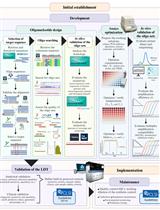
Development of a Novel Automated Workflow in Fiji ImageJ for Batch Analysis of Confocal Imaging Data to Quantify Protein Colocalization Using Manders Coefficient
基于Fiji ImageJ的全自动化流程开发:批量分析共聚焦图像数据并量化蛋白共定位的Manders系数
Screening for Streptococcus agalactiae: Development of an Automated qPCR-Based Laboratory-Developed Test Using Panther Fusion® Open AccessTM
链球菌筛查:基于Panther Fusion® Open Access™平台开发的自动化qPCR实验室自建检测方法
Pupillometry: A Simple and Automatic Way to Explore Implicit Cognitive Processing
瞳孔测量:探索内隐认知加工的简单自动方法
生物工程
A Human Cervix Chip for Preclinical Studies of Female Reproductive Biology
用于女性生殖生物学前沿研究的人源子宫颈芯片模型
癌症生物学
A Protocol for Laser-Assisted Microdissection and tRF & tiRNA Sequencing in Lung Adenocarcinoma
肺腺癌中激光显微切割与tRF/tiRNA测序操作方法
Assay for Site-Specific Homologous Recombination Activity in Adherent Cells, Suspension Cells, and Tumor Tissues
贴壁细胞、悬浮细胞及肿瘤组织中位点特异性同源重组活性的检测方法
细胞生物学
Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
小鼠咬肌肌纤维类型分析的共聚焦成像方案优化
Isolation of In Vitro Osteoblastic-Derived Matrix Vesicles by Ultracentrifugation and Cell-Free Mineralization Assay
体外成骨细胞来源基质囊泡的超速离心分离及无细胞成矿分析方法
发育生物学
Single Molecule Fluorescence In Situ Hybridization Using RNAscope to Study Hematopoietic and Vascular Interactions in the Zebrafish Embryo and Larva
利用RNAscope单分子荧光原位杂交技术研究斑马鱼胚胎和幼体中的造血与血管互作
环境生物学
Baculovirus and Plasmid Vector-Mediated Gene Transfer in Daphnia magna Cells in Primary Embryonic In Vitro Cultures in an Optimized Microenvironment: Methods and Protocols
在优化微环境中利用杆状病毒和质粒载体介导的大型蚤胚胎原代细胞基因转导:方法与方案
免疫学
Analysis of Vascular Permeability by a Modified Miles Assay
改良版Miles法分析血管通透性
A Participant-Derived Xenograft Mouse Model to Decode Autologous Mechanisms of HIV Control and Evaluate Immunotherapies
基于参与者来源的异种移植小鼠模型:解析HIV自体控制机制并评估免疫疗法
微生物学
Detection and Analysis of S-Acylated Proteins via Acyl Resin–Assisted Capture (Acyl-RAC)
利用酰基树脂辅助捕获法(Acyl-RAC)检测与分析S-酰化蛋白
Flotation Assay With Fluorescence Readout to Study Membrane Association of the Enteroviral Peripheral Membrane Protein 2C
基于荧光读数的漂浮分析法研究肠道病毒周边膜蛋白2C的膜结合特性
植物科学
Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
甜樱桃发育中花原基的淀粉酶法定量检测
Analysis of Modified Plant Metabolites Using Widely Targeted Metabolite Modificomics
基于广泛靶向代谢修饰组学的植物修饰代谢产物分析
A Detailed Guide to Recording and Analyzing Arabidopsis thaliana Leaf Surface Potential Dynamics Elicited by Mechanical Wounding
拟南芥机械损伤引发的叶片表面电位动态记录与分析详解指南
High-Performance Liquid Chromatography Quantification of Glyphosate, Aminomethylphosphonic Acid, and Ascorbate in Culture Medium and Microalgal Cells
高效液相色谱法检测培养基及微藻细胞中草甘膦、氨基甲基膦酸和抗坏血酸的含量